TBtrans#
Here is the instruction for an deeptb interface tbtrans_negf, which is convenient for users with needs to do NEGF calculation and are familiar with TBtrans.
This interface enpowers deeptb the ability to generate input files for TBtrans, containing sufficient information for TBtrans to calculate transport properties.
TBtrans info#
TBtrans (Tight-Binding transport) is a generic computer program which calculates transport and other physical quantities using the Green function formalism. It is a stand-alone program which allows extreme scale tight-binding calculations. For details, see https://www.sciencedirect.com/science/article/pii/S001046551630306X?via%3Dihub.
We design an inferface for dptb to utilize TBtrans as a tool for transport calculation. The interface would prepare tbtrans input files (.nc) by extracting necessary information from deeptb-negf input files and predicting hamiltonian with deeptb model.
Users only need to prepare their own RUN.fdf (TBtrans input setting files) to satisfy various functions in TBtrans, although we have provided an example input in DeePTB/examples/tbtrans_hBN/tbtrans_input/RUN_tbtrans.fdf.
To run the Interface, users need install sisl package(https://zerothi.github.io/sisl/index.html), which is a postprocess python code for siesta.
Attention:
TBtrans would not automatically run. This interface just prepares input files for you!
If you are interested in Runing TBtrans, please install it firstly.
Currently,
tbtrans_negfonly support 2-terminal transport.
Input files for tbtrans_negf#
To run tbtrans_negf, only 3 files user should prepare: structure, deeptb model and input setting.
structure#
tbtrans_negfsupportextxyzand.vaspformat.Following the same convention as dptb-negf, the transport direction should be the z-direction.
deeptb model#
tbtrans_negfwould use this model to generate the Hamiltonian and related info for later transport calculation.
input setting#
input settingcontains information of the region definition and periodic conditions. An example file isDeePTB/examples/tbtrans_hBN/negf_tbt.json, from which this interface would automatically extract necessary information for TBtrans calculation.Users could directly reuse setting for dptb-negf as an setting for dptb-tbtrans. The only thing users need to do is give
tasklabel a new string: renamenegfastbtrans_negf.
RUN#
The interface tbtrans_negf has merged into DeePTB Shell Command and
Take DeePTB/examples/tbtrans_hBN/negf_tbt.json as an example.
cd DeePTB/examples/tbtrans_hBN/tbtrans_input/
dptb run -sk negf_tbt.json -i ./data/model/latest_nnsk_b3.600_c3.600_w0.300.pth -o tbtrans_input
As DeePTB/examples/tbtrans_hBN/data/model/latest_nnsk_b3.600_c3.600_w0.300.pth is nnsk model, we need to add -sk here to the command line. Then the input files for TBtrans would store in tbtrans_input.
Output#
structure files
lead_L_tbtrans.xyz, lead_R_tbtrans.xyz: lead region of the whole structre, being easy for users to check and visulize their definiton of leads.
srtuctre_tbtrans.vasp, structure_tbtrans.xyz: the whole structure, being easy for visulization in vasp format.
nc files
lead_L.nc, lead_R.nc: Hamiltonian and overlap matrix of lead_L and lead_R, necessary for TBtrans to calculate lead self energy.
structure.nc: Hamiltonian and overlap matrix of the whole structure
To run TBtrans successfully, users should know the basic principles in NEGF and prepare their own setting files (usually named
RUN_tbtrans.fdf) for TBtrans. Here we provide an simple exampleDeePTB/examples/tbtrans_hBN/tbtrans_input/RUN_tbtrans.fdf.
Example#
We design an example of tbtrans_negf for hBN in DeePTB/examples/tbtrans_hBN, and DeePTB/examples/tbtrans_hBN/tbtrans_hBN_show.ipynb shows the results of TBtrans transmission calculation. The detailed information and results are concluded in DeePTB/examples/tbtrans_hBN/tbtrans_hBN_show.ipynb.
Firstly we calculate the \(\Gamma\) point transmission, which evidently shows the band gap in the middle.
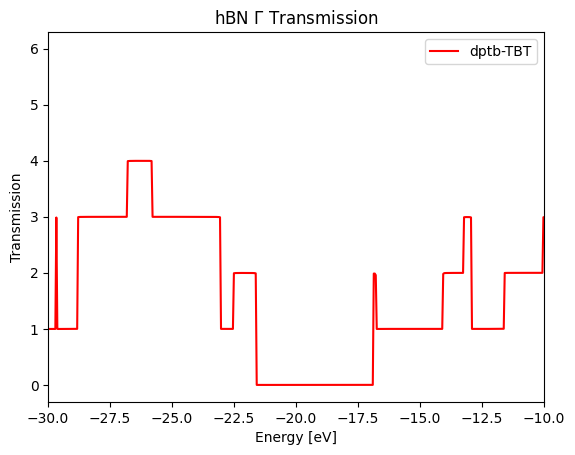
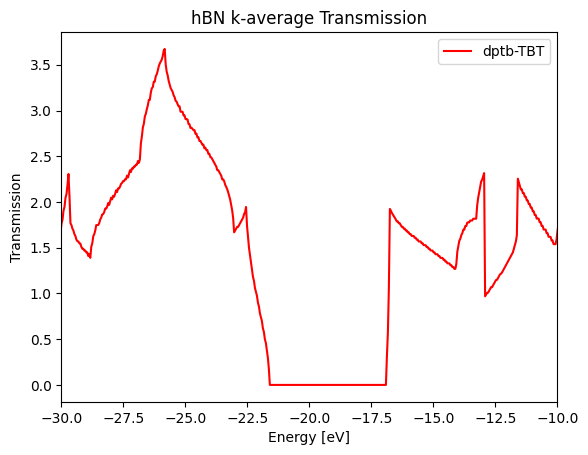
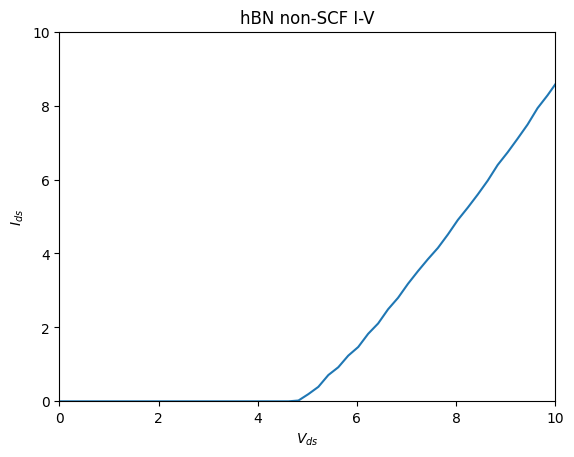
Combining this example would be the most efficient way to master tbtrans_negf interface.
