DFTB SK Files to DeePTB Model#
This example demonstrates how to build a DeePTB model from DFTB SK files. The DFTB SK files are generated by the DFTB+ code.
Example: train hBN model with DFTB SK files#
Step 1: Download DFTB SK Files#
you can download the skfiles from the dftb.org website. Here we provide the some sk files in the folder examples/hBN_dftb/slakos.
examples/hBN_dftb/slakos
├── B-B.skf
├── B-N.skf
├── C-C.skf
├── C-H.skf
├── H-C.skf
├── H-H.skf
├── N-B.skf
├── N-N.skf
└── Si-Si.skf
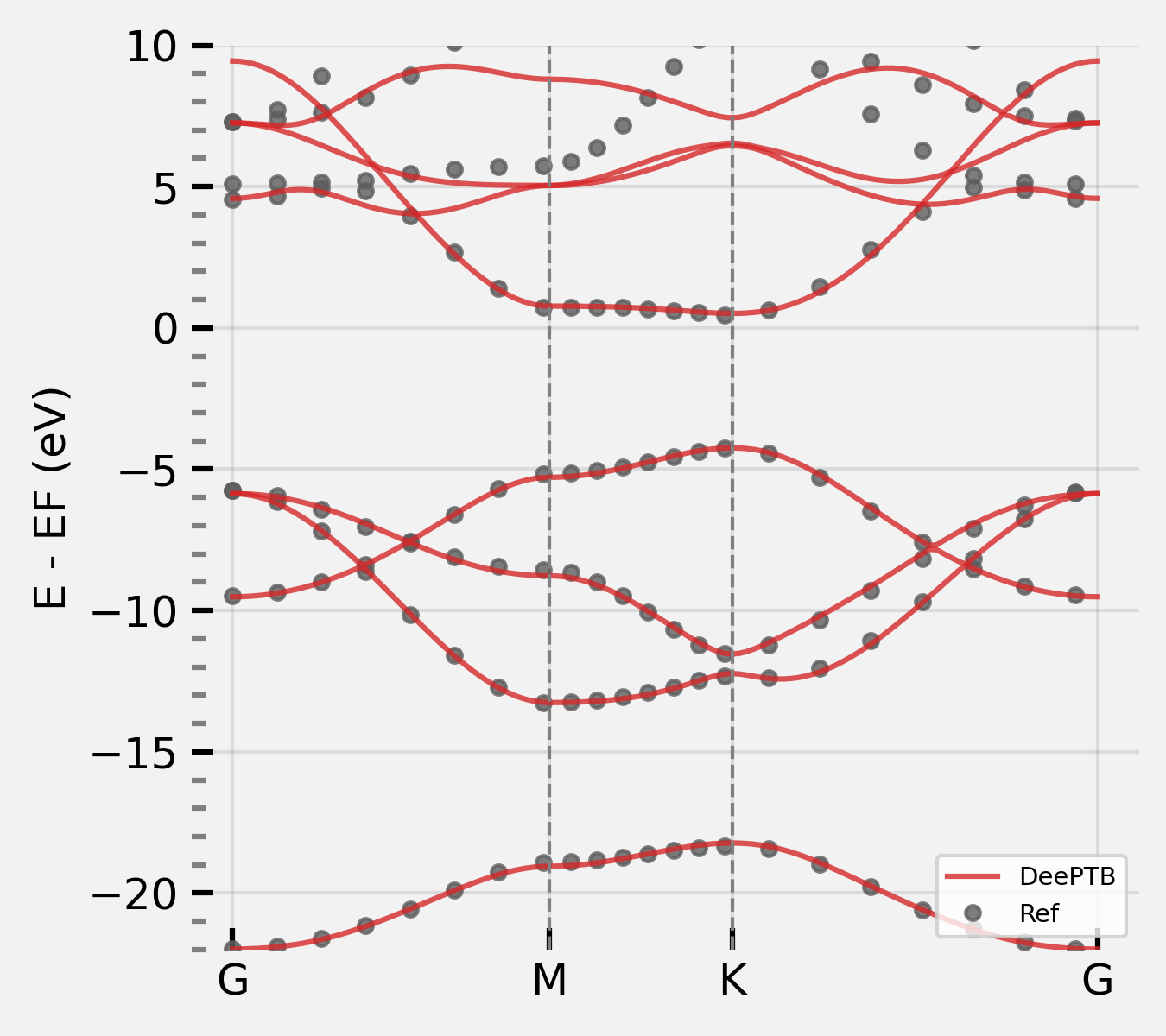
Step 2: Load DFTB skfiles to in DeePTB to plot band structure:#
cd examples/hBN_dftb
from dptb.nn.dftbsk import DFTBSK
from ase.io import read
from dptb.data import AtomicData, AtomicDataDict
from dptb.utils.tools import j_loader
import torch
from dptb.postprocess.bandstructure.band import Band
skdata = './slakos'
basis = {'B':['2s','2p'],"N":["2s","2p"]}
model = DFTBSK(basis=basis, skdata=skdata,overlap=True)
task_options={
"task": "band",
"kline_type":"abacus",
"kpath":[
[0, 0, 0, 50],
[0.5, 0, 0, 50],
[0.3333333, 0.3333333, 0, 50],
[0, 0, 0, 1]
],
"nel_atom":{"N":5,"B":3},
"klabels":["G", "M", "K", "G"],
"E_fermi":-12.798759460449219,
"emin":-25,
"emax":15,
"ref_band": "./data/kpath.0/eigenvalues.npy"
}
kpath_kwargs = task_options
stru_data = "./data/struct.vasp"
AtomicData_options = {"r_max": 5.0, "oer_max":1.6, "pbc": True}
structase = read(stru_data)
bcal = Band(model=model,
use_gui=False,
results_path="./",
device=model.device)
eigenstatus = bcal.get_bands(data=stru_data,
kpath_kwargs=kpath_kwargs,
AtomicData_options=AtomicData_options)
bcal.band_plot(ref_band = kpath_kwargs["ref_band"],
E_fermi = -2,
emin = -25,
emax = 15)
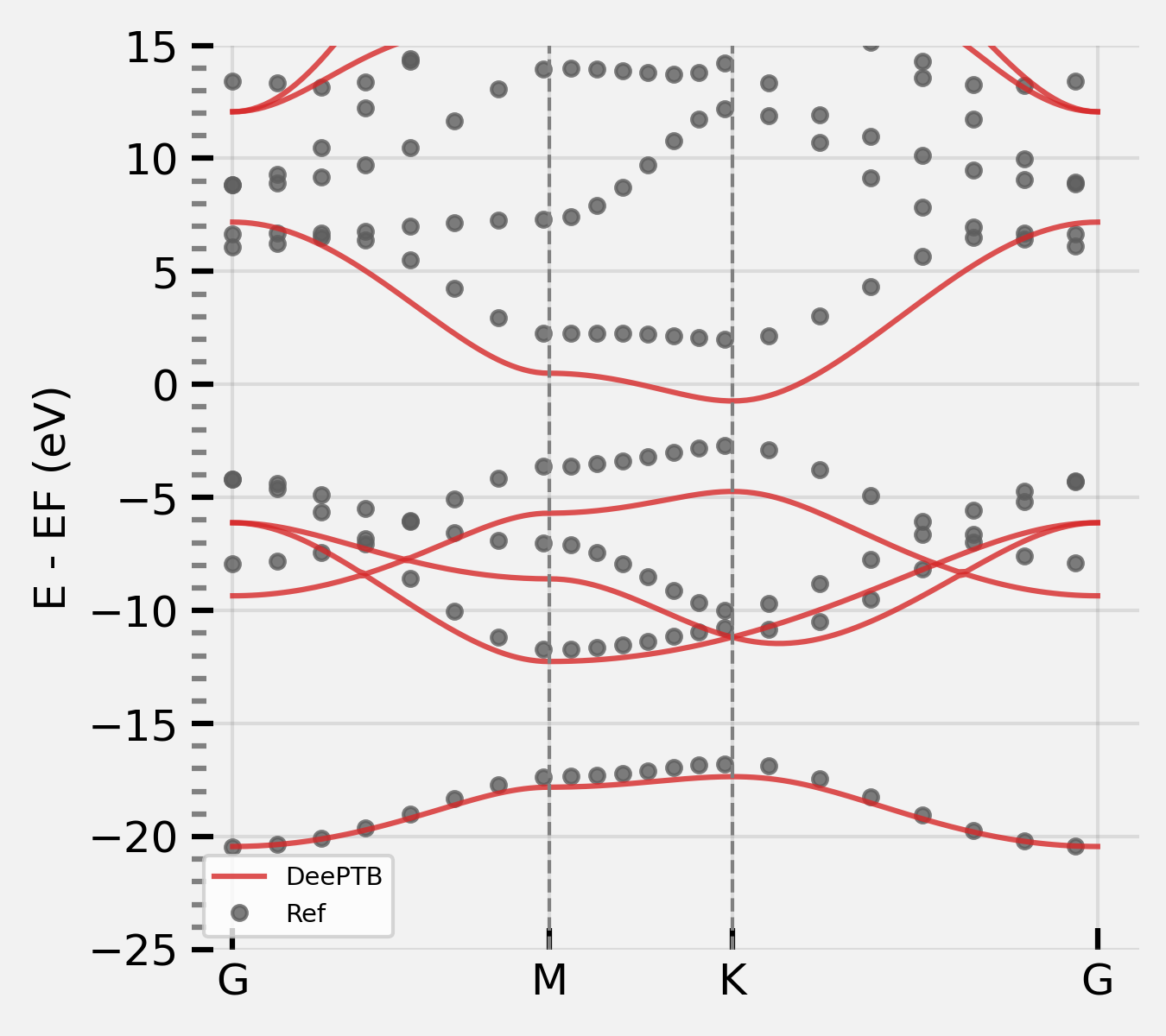
Step 3: Use SK params from DFTB skfiles to train DeePTB nnsk model#
use the input.json in the following:
{
"common_options": {
"basis": {
"B": ["2s", "2p"],
"N": ["2s", "2p"]
},
"device": "cpu",
"dtype": "float32",
"overlap": false,
"seed": 42
},
"train_options": {
"num_epoch": 500,
"batch_size": 1,
"optimizer": {
"lr": 0.01,
"type": "Adam"
},
"lr_scheduler": {
"type": "exp",
"gamma": 0.999
},
"loss_options":{
"train": {"method": "skints", "skdata":"./slakos"}
},
"save_freq": 50,
"validation_freq": 10,
"display_freq": 10
},
"model_options": {
"nnsk": {
"onsite": {"method": "uniform"},
"hopping": {"method": "powerlaw", "rs":4.5, "w": 0.2},
"soc":{},
"freeze": false,
"push":false
}
},
"data_options": {
"train": {
"root": "./data/",
"prefix": "kpath",
"get_eigenvalues": false
}
}
}
And then run the following command:
dptb train input_nnsk_skints.json -o ./nnskint
Then you can get the model in nnskint/checkpoint;
note The overlap in common_options is set to false. The overlap in DFTB sk files will not be used.
We can see the band structure using the following code:
from dptb.nn.build import build_model
from dptb.utils.tools import j_loader
from dptb.postprocess.bandstructure.band import Band
model = build_model(checkpoint="./nnskint/checkpoint/nnsk.best.pth")
task_options={
"task": "band",
"kline_type":"abacus",
"kpath":[
[0, 0, 0, 50],
[0.5, 0, 0, 50],
[0.3333333, 0.3333333, 0, 50],
[0, 0, 0, 1]
],
"nel_atom":{"N":5,"B":3},
"klabels":["G", "M", "K", "G"],
"E_fermi":-12.798759460449219,
"emin":-25,
"emax":15,
"ref_band": "./data/kpath.0/eigenvalues.npy"
}
kpath_kwargs = task_options
stru_data = "./data/struct.vasp"
AtomicData_options = {"r_max": 5.0,"er_max": 3.5, "oer_max":1.6, "pbc": True}
bcal = Band(model=model,
use_gui=False,
results_path='./',
device=model.device)
eigenstatus = bcal.get_bands(data=stru_data,
kpath_kwargs=kpath_kwargs,
AtomicData_options=AtomicData_options)
bcal.band_plot(ref_band = kpath_kwargs["ref_band"],
E_fermi = -5,
emin = -22,
emax = 10)
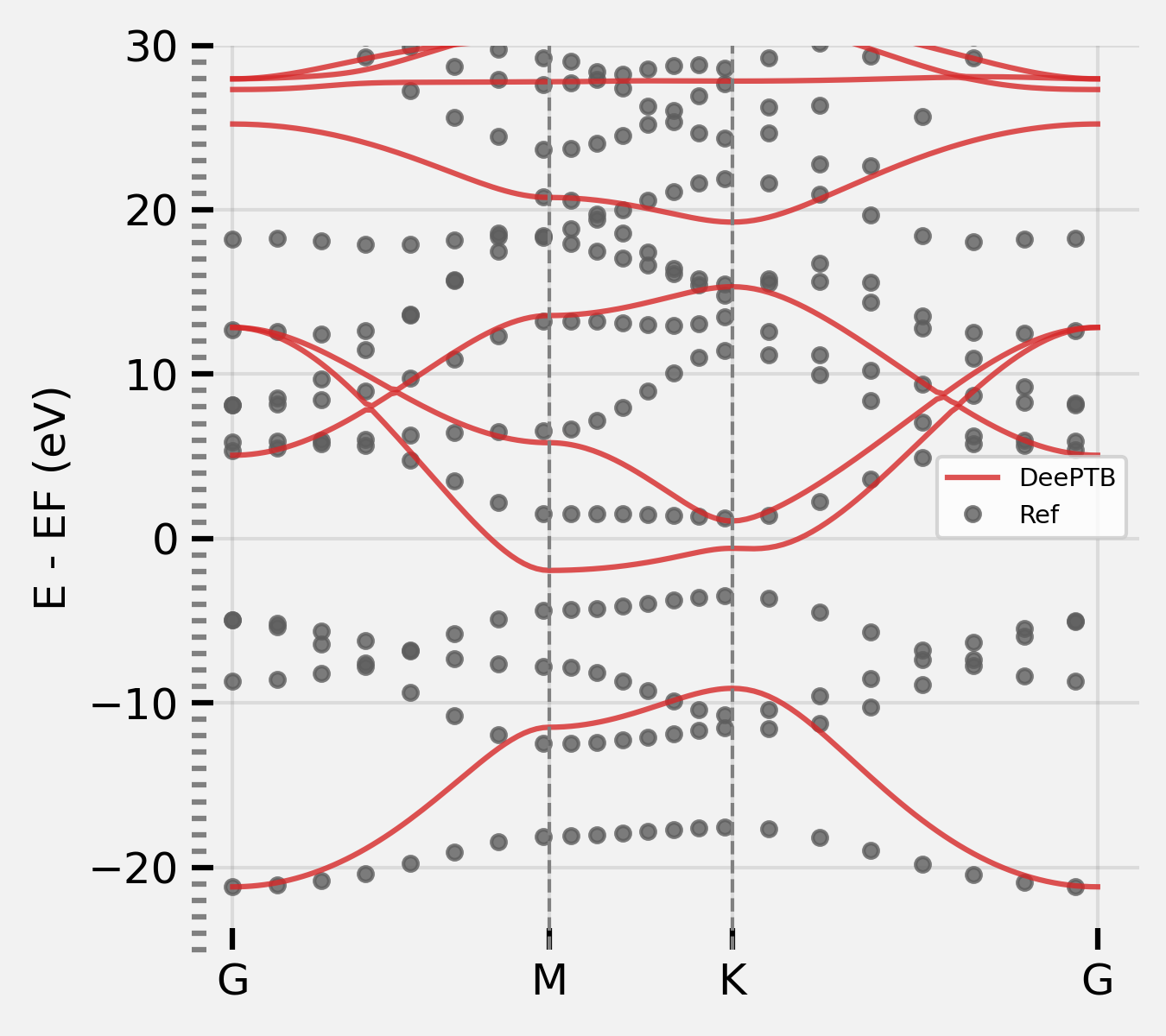
NOTE: By setting the “overlap”: true, and retrain the model, deeptb can get the overlap back.
dptb train input_nnsk_skints.json -o ./nnskint_overlap
from dptb.nn.build import build_model
from dptb.utils.tools import j_loader
from dptb.postprocess.bandstructure.band import Band
model = build_model(checkpoint="./nnskint_overlap/checkpoint/nnsk.best.pth")
task_options={
"task": "band",
"kline_type":"abacus",
"kpath":[
[0, 0, 0, 50],
[0.5, 0, 0, 50],
[0.3333333, 0.3333333, 0, 50],
[0, 0, 0, 1]
],
"nel_atom":{"N":5,"B":3},
"klabels":["G", "M", "K", "G"],
"E_fermi":-12.798759460449219,
"emin":-25,
"emax":15,
"ref_band": "./data/kpath.0/eigenvalues.npy"
}
kpath_kwargs = task_options
stru_data = "./data/struct.vasp"
AtomicData_options = {"r_max": 5.0,"er_max": 3.5, "oer_max":1.6, "pbc": True}
bcal = Band(model=model,
use_gui=False,
results_path='./',
device=model.device)
eigenstatus = bcal.get_bands(data=stru_data,
kpath_kwargs=kpath_kwargs,
AtomicData_options=AtomicData_options)
bcal.band_plot(ref_band = kpath_kwargs["ref_band"],
E_fermi = -5,
emin = -22,
emax = 10)
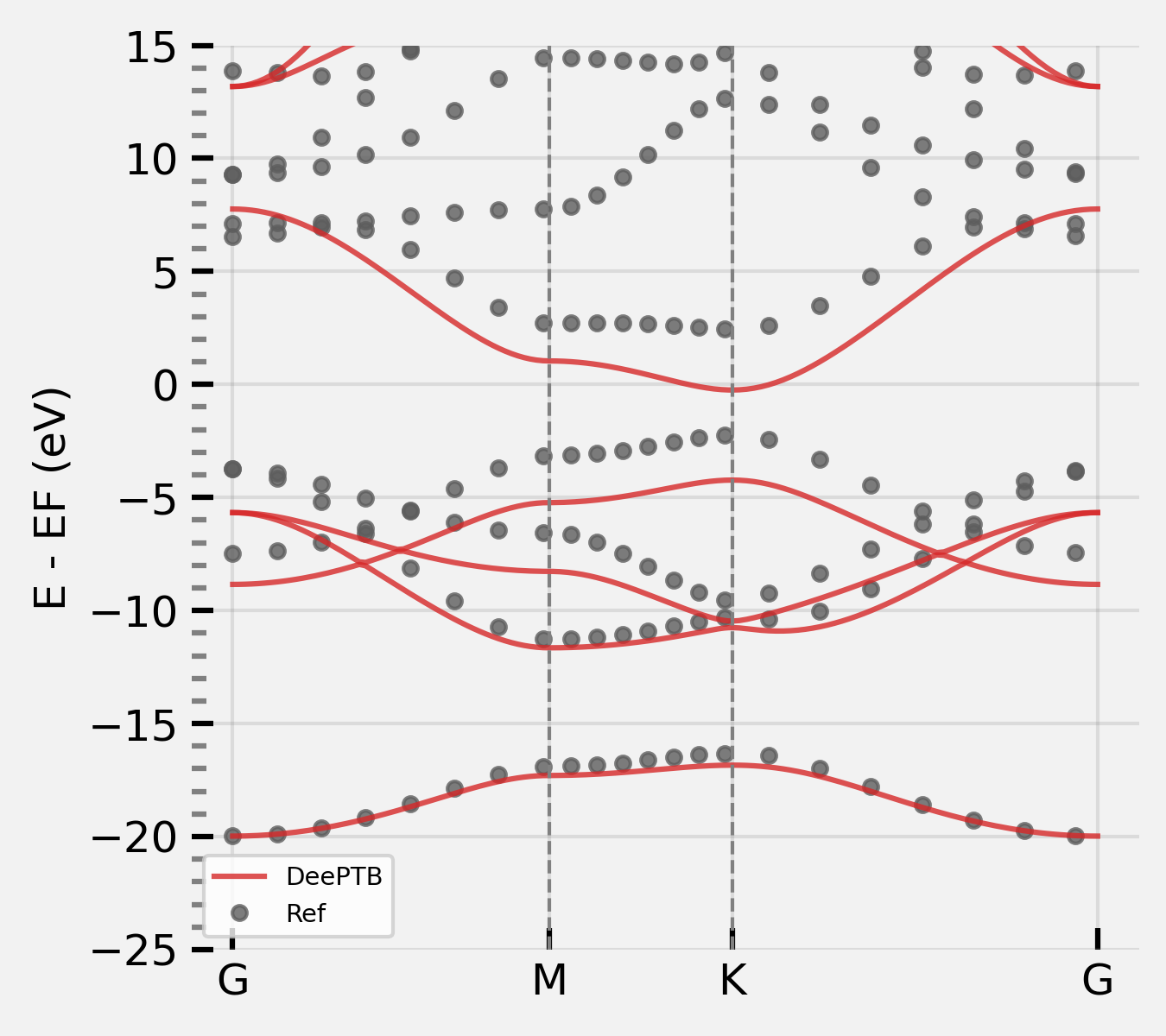
Step4: load the previous trained model to further train the model use eigenvalues#
The input json looks like:
{
"common_options": {
"basis": {
"B": ["2s", "2p"],
"N": ["2s", "2p"]
},
"device": "cpu",
"dtype": "float32",
"overlap": false,
"seed": 42
},
"train_options": {
"num_epoch": 500,
"batch_size": 1,
"optimizer": {
"lr": 0.01,
"type": "Adam"
},
"lr_scheduler": {
"type": "exp",
"gamma": 0.999
},
"loss_options":{
"train": {"method": "eigvals"}
},
"save_freq": 50,
"validation_freq": 10,
"display_freq": 10
},
"model_options": {
"nnsk": {
"onsite": {"method": "uniform"},
"hopping": {"method": "powerlaw", "rs":4.5, "w": 0.2},
"soc":{},
"freeze": false,
"push":false
}
},
"data_options": {
"train": {
"root": "./data/",
"prefix": "kpath",
"get_eigenvalues": true
}
}
}
None: we want have the orthogonal TB model. The overlap in common_options is set to false. The overlap in DFTB sk files will not be used.
dptb train input_nnsk_eigvals.json -i nnskint/checkpoint/nnsk.best.pth -o ./nnsk_eigvals
Show the bands:
from dptb.nn.build import build_model
from dptb.utils.tools import j_loader
from dptb.postprocess.bandstructure.band import Band
model = build_model(checkpoint="./nnsk_eigvals/checkpoint/nnsk.ep500.pth")
jdata = j_loader("./run/band.json")
results_path = "./band_plot"
kpath_kwargs = jdata["task_options"]
stru_data = "./data/struct.vasp"
AtomicData_options = {"r_max": 5.0,"er_max": 3.5, "oer_max":1.6, "pbc": True}
bcal = Band(model=model,
use_gui=False,
results_path='./',
device=model.device)
eigenstatus = bcal.get_bands(data=stru_data,
kpath_kwargs=kpath_kwargs,
AtomicData_options=AtomicData_options)
bcal.band_plot(ref_band = kpath_kwargs["ref_band"],
E_fermi = -5,
emin = -22,
emax = 10)